

2025

62. A Robust Ensemble Machine Learning Approach for Inhibitor Discovery: Case Study of HIV-1 NNRTI and Validation Using MD Simulation
A. Shree, P. Pani, M. K. Rana


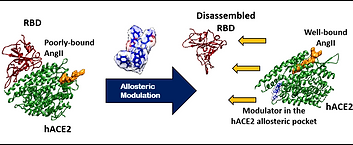
61. Modulating functional allostery of the host-cell receptor protein hACE2 to inhibit viral entry of SARS-CoV-2
P. Pani, S. K. Panda, M. K. Rana

60. Unleashing the Potential of a Zr-MOF for Selective Amine and Imine Synthesis: An Efficient Heterogeneous Catalyst
D. Singha, J. Panda, M. K. Rana


59. In silico evaluation of S-adenosyl-L-homocysteine analogs as inhibitors of nsp14-viral cap N7 methyltranferase and PLpro of SARS-CoV-2: synthesis, molecular docking, physicochemical data, ADMET and molecular dynamics simulations studies
R. Srivastava, S. K. Panda, P. S. S. Gupta, A. Chaudhary, F. Naaz, A. K. Yadav, N. K. Ram, M. K. Rana, R. K. Singh, R. Srivastava
Journal of Biomolecular Structure and Dynamics, 43(7), 3258–3275.


58. In silico evidence of monkeypox F14 as a ligand for the human TLR1/2 dimer
A. Chakraborty, N. C. Das, P. S. S. Gupta, S. K. Panda, M. K. Rana, S. R. Bonam, J. Bayry, S. Mukherjee


57. Rapid adsorption of industrial cationic dye pollutant using base-activated rice straw biochar: performance, isotherm, kinetic and thermodynamic evaluation
S. Senapati, J. Giri, L. Mallick, D, Singha, T. Bastia, P. Rath, M. K. Rana, A. K. Panda


56. Investigation of metal–organic frameworks and fluorocarbon refrigerants promising for adsorption cooling systems
D. Singha, D. Mohapatra, M. K. Rana
Phys. Chem. Chem. Phys., 2025,27, 11530-1154 (Cover Art Publication)


55. Computational Assessment of Clinical Drugs against SARS-CoV-2: Foreseeing Molecular Mechanisms and Potent Mpro Inhibitors
S. K. Panda, P. Pani, P. S. S. Gupta, N. C. Mahanandia, M. K. Rana

2024

54. Uncovering the Latent Responsiveness of the Water-Stable, Luminescent Zn-BDC MOF with Distinct Morphology for Advanced Sensing of Nitro Explosives and Ions
D. Singha, J. Panda, M. K. Rana
The Journal of Physical Chemistry C, 2024, 128, 44, 19013–19023


53. Immune targeting of filarial glutaredoxin through a multi-epitope peptide-based vaccine: A reverse vaccinology approach
N. C. Das, S. Gorai, P. S. S. Gupta, S. K. Panda, M. K. Rana, S. Mukherjee


52. Critical assessment of interactions between ct-DNA and choline-based magnetic ionic liquids: evidences of compaction
K. D. Tulsiyan, S. K. Panda, M. K. Rana, H. S. Biswal


51. Can Duvelisib and Eganelisib work for both cancer and COVID-19? Molecular-level insights from MD simulations and enhanced sampling
S. K. Panda, S. Karmakar, P. S. S. Gupta, M. K. Rana

2023


50. Unmasking an Allosteric Binding Site of the Papain-like Protease in SARS-CoV-2: Molecular Dynamics Simulations of Corticosteroids.
S. K. Panda, P. S. S. Gupta, S. Karmakar, S. Biswal, N. C. Mahanandia, M. K. Rana
J. Phys. Chem. Lett. 2023, 14, 45, 10278–10284 (Cover Art Publication)

49. Post-synthetic Modification of Zr-based Metal-organic Frameworks with Imidazole: Variable Optical Behavior and Sensing.
D. P. Biswal, D. Singha, J. Panda, M. K. Rana


48. A microporous water stable MOF for consistent and selective C₂H₂/C₂H₄ separation.
A. Pal, S. C. Pal, H. Cui, R. B. Lin, D. Singha, M. K. Rana, B. Chen, M. C. Das


47. Exploring antiviral potency of N-1 substituted pyrimidines against HIV-1 and other DNA/RNA viruses: Design, synthesis, characterization, ADMET analysis, docking, molecular dynamics and biological activity.
R. Srivastava, S. K. Gupta, F. Naaz, P. S. S. Gupta, M. Yadav, V. K. Singh, S. K. Panda, S. Biswal, M. K. Rana, S. K. Gupta, D. Schols, R. K. Singh


46. Design, synthesis, antimicrobial activity, DFT, and molecular docking studies of pyridine‐pyrazole‐based dihydro‐1, 3, 4‐oxadiazoles against various bacterial and fungal targets.
N. Desai, D. Jadeja, J. Monapara, S. K. Panda, M. K. Rana, B. Dave
Journal of Biochemical and Molecular Toxicology, 2023, e23377.


45. Identification and investigation of a cryptic binding pocket of the P37 envelope protein of monkeypox virus by molecular dynamics simulations.
P. S. S. Gupta, S. K. Panda, A. K. Nayak, M. K. Rana
The Journal of Physical Chemistry Letters, 2023, 14(13), 3230-3235.


44. A pH‐Activatable Prodrug and Metal Prodrug Conjugate of Gossypol: Synthesis, Emergent Photophysical, Nanoscopic, Computational, and in‐Vitro Cellular Studies.
M. Biswas, Kajol, D. Singha, M. K. Rana, R. K. Pathak.


43. Insights into the binding mechanism of ascorbic acid and violaxanthin with violaxanthin de-epoxidase (VDE) and chlorophycean violaxanthin de-epoxidase (CVDE) enzymes.
S. Biswal, P. S. S. Gupta, S. K. Panda, H. R. Bhat, M. K. Rana


42. Cell surface fibroblast activation protein-2 (Fap2) of fusobacterium nucleatum as a vaccine candidate for therapeutic intervention of human colorectal cancer: an immunoinformatics approach.
S. Padma, R. Patra, P. S. S. Gupta, S. K. Panda, M. K. Rana, S. Mukherjee


41. Reverse vaccinology assisted design of a novel multi-epitope vaccine to target Wuchereria bancrofti cystatin: An immunoinformatics approach.
N. C. Das, P. S. S. Gupta, S. K. Panda, M. K. Rana, S. Mukherjee


40. Sensing cyclosarin (a chemical warfare agent) by Cucurbit [n] urils: A DFT/TD-DFT study.
H. R. Bhat, M. K. Rana, A. A. Dar

2022

39. Potential targets of severe acute respiratory syndrome coronavirus 2 of clinical drug fluvoxamine: Docking and molecular dynamics studies to elucidate viral action.
S. K. Panda, P. S. S. Gupta, M. K. Rana


38. α-graphyne as a promising anode material for Na-ion batteries: a first-principles study.
T. Singh, J. R. Choudhuri, M. K. Rana


37. Hydroxyalkynyl uracil derivatives as NNRTIs against HIV-1: in silico predictions, synthesis, docking and molecular dynamics simulation studies.
M. Yadav, R. Srivastava, F. Naaz, P. S. S. Gupta, S. K. Panda, M. K. Rana, R. K. Singh


36. Triazine skeletal covalent organic frameworks: A versatile highly positive surface potential triboelectric layer for energy harvesting and self-powered applications.
S. Hajra, J. Panda, J. Swain, H. G. Kim, M. Sahu, M. K. Rana, R. Samantaray, H. J. Kim, R. Sahu


35. Experimental and DFT study of transition metal doping in a Zn-BDC MOF to improve electrical and visible light absorption properties.
J. Panda, D. Singha, P. K. Panda, B. C. Tripathy, M. K. Rana
The Journal of Physical Chemistry C, 2022, 126(30), 12348-12360.


34. Inhibitors of Heptosyltransferase I to prevent heptose transfer against antibiotic resistance of E. coli: Energetics and stability analysis by DFT and molecular dynamics.
U. Bhattacharya, S. K. Panda, P. S. S. Gupta, M. K. Rana


33. Designing AbhiSCoVac-A single potential vaccine for all ‘corona culprits’: Immunoinformatics and immune simulation approaches.
A. Choudhury, P. S. S. Gupta, S. K. Panda, M. K. Rana, S. Mukherjee


32. Repurposing of FDA-approved drugs as potential inhibitors of the SARS-CoV-2 main protease: Molecular insights into improved therapeutic discovery.
A. K. Ray, P. S. S. Gupta, S. K. Panda, S. Biswal, U. Bhattacharya, M. K. Rana


31. Designing efficient multi-epitope peptide-based vaccine by targeting the antioxidant thioredoxin of bancroftian filarial parasite.
S. Gorai, N. C. Das, P. S. S. Gupta, S. K. Panda, M. K. Rana, & S. Mukherjee.

2021

30. Inhibitors of Plasmepsin X Plasmodium falciparum: Structure-based pharmacophore generation and molecular dynamics simulation.
S. K. Panda, S. Saxena, P. S. S. Gupta, M. K. Rana


29. In-silico evidences on filarial cystatin as a putative ligand of human TLR4.
N. C. Das, P. S. S. Gupta, S. Biswal, R. Patra, M. K. Rana, S Mukherjee
Journal of Biomolecular Structure and Dynamics, 2021, 40(19), 8808-8824.


28. Templated growth of polyaniline for enhanced gas sensing response in flexible sensors: experiments and simulations.
M. Sinha, M. K. Rana, S. Panda


27. ACE-2-derived biomimetic peptides for the inhibition of spike protein of SARS-CoV-2.
S. K. Panda, P. S. S. Gupta, S. Biswal, A. K. Ray, M. K. Rana


26. Design, synthesis, biological evaluation and molecular docking study of novel hybrid of pyrazole and benzimidazoles.
N. C. Desai, D. D. Pandya, D. J. Jadeja, S. K. Panda, M. K. Rana


25. Designing of a novel multi-epitope peptide based vaccine against Brugia malayi: An in silico approach.
N. C. Das, R. Patra, P. S. S. Gupta, P. Ghosh, U. Bhattacharya, M. K. Rana, S. Mukherjee
2020

24. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin.
P. S. S. Gupta, S. Biswal, S. K. Panda, A. K. Ray, M. K. Rana.
Journal of Biomolecular Structure and Dynamics, 2020, 40(5), 2217-2226.


23. Computer-aided discovery of bis-indole derivatives as multi-target drugs against cancer and bacterial infections: DFT, docking, virtual screening, and molecular dynamics studies.
P. S. S. Gupta, H. R. Bhat. S. Biswal, M. K. Rana.


22. Alkylated benzimidazoles: Design, synthesis, docking, DFT analysis, ADMET property, molecular dynamics and activity against HIV and YFV.
R. Srivastava, S. K. Gupta, F. Naaz, P. S. S. Gupta, M. Yadav, V. K. Singh, A. Singh, M. K. Rana, S. K. Gupta, D. Schols, R. K. Singh.


21. Ivermectin, famotidine, and doxycycline: a suggested combinatorial therapeutic for the treatment of COVID-19.
P. S. S. Gupta & M. K. Rana.
ACS Pharmacology & Translational Science, 2020, 3(5), 1037-1038.


20. Screening and molecular characterization of lethal mutations of human homogentisate 1, 2 dioxigenase.
P. S. S. Gupta, R. N. U. Islam, S. Banerjee, A. Nayek, M. K. Rana, A. K. Bandyopadhyay.
Journal of Biomolecular Structure and Dynamics, 2020, 39(5), 1661-1671.

19. A “thermodynamically stable” 2D nickel metal–organic framework over a wide pH range with scalable preparation for efficient C2s over C1 hydrocarbon separations.
R. Sahoo, S. Chand, M. Mondal, A. Pal, S. C. Pal, M. K. Rana, M. C. Das.


18. Anti-HIV potential of diarylpyrimidine derivatives as non-nucleoside reverse transcriptase inhibitors: Design, synthesis, docking, TOPKAT analysis and molecular dynamics simulations.
V. K. Singh, R. Srivastava, P. S. S. Gupta, F. Naaz, H. Chaurasia, R. Mishra, M. K. Rana, R. K. Singh.
Journal of Biomolecular Structure and Dynamics, 2020, 39(7), 2430-2446.

17. Binding insight of clinically oriented drug famotidine with the identified potential target of SARS-CoV-2.
P. S. S. Gupta, S. Biswal, D. Singha, M. K. Rana.
Journal of Biomolecular Structure and Dynamics, 2020, 39(14), 5327-5333.

2019

16. Exploring Bikaverin as Metal Ion Biosensor: A Computational Approach.
Z. Hussain, H. R. Bhat, T. Naqvi, M. K. Rana, M. A. Rizvi.



15. Anion sensing by novel triarylboranes containing boraanthracene: DFT functional assessment, selective interactions, and mechanism demonstration.
H. R. Bhat, P. S. S. Gupta, S. Biswal, M. K. Rana.

Before Joining IISER
14. Gas sensing behavior of metal-phthalocyanines: Effects of electronic structure on sensitivity.
Rana, M. K., Sinha, M., & Panda, S.
13. Solvation of narrow pores of graphene-like plates in simple dipolar liquids: Wetting and dewetting behavior and solvent dynamics for varying pore width and solute–solvent interaction.
Rana, M. K., & Chandra, A.
12. Predicting methane storage in open-metal-site metal–organic frameworks.
Koh, H. S., Rana, M. K., Wong-Foy, A. G., & Siegel, D. J.
The Journal of Physical Chemistry C, 2015, 119(24), 13451-13458.
11. Wetting behavior of nonpolar nanotubes in simple dipolar liquids for varying nanotube diameter and solute-solvent interactions.
Rana, M. K., & Chandra, A.
10. Methane storage in metal-substituted metal–organic frameworks: thermodynamics, usable capacity, and the impact of enhanced binding sites.
Rana, M. K., Koh, H. S., Zuberi, H., & Siegel, D. J.
The Journal of Physical Chemistry C, 2014, 118(6), 2929-2942.
9. Simulation study of CO2 adsorption properties in small Zeolite Imidazolate Frameworks.
Rana, M. K., Suffritti, G. B., Demontis, P., & Masia, M.
8. Thermodynamic screening of metal-substituted MOFs for carbon capture.
Koh, H. S., Rana, M. K., Hwang, J., & Siegel, D. J.
Physical Chemistry Chemical Physics, 2013, 15(13), 4573-4581.
7. Ab initio and classical molecular dynamics studies of the structural and dynamical behavior of water near a hydrophobic graphene sheet.
Rana, M. K., & Chandra, A.
6. Solvation structure of nanoscopic hydrophobic solutes in supercritical water: Results for varying thickness of hydrophobic walls, solute–solvent interaction and solvent density.
Rana, M. K., & Chandra, A.
5. Solvation of fullerene and fulleride ion in liquid ammonia: Structure and dynamics of the solvation shells.
Rana, M. K., & Chandra, A.
4. Comparing van der Waals density functionals for CO2 adsorption in metal organic frameworks.
Rana, M. K., Koh, H. S., Hwang, J., & Siegel, D. J.
The Journal of Physical Chemistry C, 2012, 116(32), 16957-16968.
3. Estimation of partial charges in small zeolite imidazolate frameworks from density functional theory calculations.
Rana, M. K., Pazzona, F. G., Suffritti, G. B., Demontis, P., & Masia, M.
Journal of Chemical Theory and Computation, 2011, 7(6), 1575-1582.
2. Water structure near single and multi-layer nanoscopic hydrophobic plates of varying separation and interaction potentials.
Rana, M. K., & Chandra, A.
1. Filled and empty states of carbon nanotubes in water: Dependence on nanotube diameter, wall thickness and dispersion interactions.
Rana, M. K., & Chandra, A.
